Genetic Testing for Acute Myeloid Leukemia - CAM 306
Description
Acute myeloid leukemia (AML) is characterized by large numbers of abnormal, immature myeloid cells in the bone marrow and peripheral blood resulting from genetic changes in hematopoietic precursor cells which disrupt normal hematopoietic growth and differentiation (Stock, 2020).
Policy
-
Genetic testing for FLT3 internal tandem duplication and tyrosine kinase domain mutations (ITD and TKD), IDH1, IDH2, TET2, WT1, DNMT3A, ASXL1 and/or TP53 is considered MEDICALLY NECESSARY in adult and pediatric patients with suspected or confirmed acute myeloid leukemia (AML) of any type for prognostic and/or therapeutic purposes.
-
Genetic testing for KIT mutations is considered MEDICALLY NECESSARY for adult and pediatric patients with confirmed core-binding factor (CBF) AML (AML with t(8;21)(q22;q22.1); RUNX1-RUNX1T1 or inv(16)(p13.1q22) /t(16;16)(p13.1;q22); CBFB-MYH11).
-
Genetic testing for NPM1, CEBPA, and RUNX1 mutations is considered MEDICALLY NECESSARY for patients other than those with confirmed core binding factor AML or AML with myelodysplasia-related cytogenetic abnormalities.
The following does not meet coverage criteria due to a lack of available published scientific literature confirming that the test(s) is/are required and beneficial for the diagnosis and treatment of a patient’s illness.
- The use of global/gene specific methylation, micro RNA (miRNA) expression, or gene expression analysis for diagnosis or prognosis in patients with confirmed acute leukemia is investigational/unproven therefore considered NOT MEDICALLY NECESSARY.
Policy Guidelines
Genetic testing for cytogenetically normal acute myeloid leukemia is intended to guide management decisions in patients who would receive treatment other than low-dose chemotherapy or best supportive care.
Rationale
Acute myeloid leukemia (AML) is the most common acute leukemia in adults (80%), with a median age at diagnosis of 65 years. An AML diagnosis in adults is generally associated with a poor prognosis. This disease is much less common in children younger than 10 years of age, as less than 10% of acute leukemias are diagnosed as AML in this age group (Schiffer & Gurbaxani, 2022; Siegel et al., 2017; Yamamoto & Goodman, 2008).
A clinical presentation of AML includes symptoms related to complications of pancytopenia, including weakness and fatigability, infections of variable severity, and hemorrhagic findings (Schiffer & Gurbaxani, 2022). Analysis of gene sequencing of AML cases generally reveal more than 10 significant gene mutations; many of which are thought to participate in leukemogenesis (CGARN, 2013). The most common gene mutations are as follows: FLT3 (28%), NPM1 (27%), DNMT3A (26%), IDH1 or IDH2 (20%), NRAS or KRAS (12%), RUNX1 (10%), TET2 (8%), TP53 (8%), CEBPA (6%), and WT1 (6%). Mutations impacting signal activation are frequent (60% of cases); the most common of which are mutations in FLT3 (Stock, 2020).
FMS-like tyrosine kinase 3 (FLT3) is a transmembrane tyrosine kinase receptor that stimulates cell proliferation upon activation. Both internal tandem duplications (ITDs) of different lengths and point mutations in the activating loop of the kinase domain result in ligand-independent activation of the FLT3 receptor and a proliferative signal. A FLT3-ITD mutation has been shown to have a poor prognosis in contrast to FTL3 point mutations in the activation loop of the kinase domain. Higher ratios of mutated alleles compared to wild-type alleles confer worse prognoses (Schiffer, 2021).
The second most common mutation in AML (27%) is of nucleophosmin (NPM1), a ubiquitously expressed phosphoprotein that normally shuttles between the nucleus and cytoplasm (Falini et al., 2005; Stock, 2020). NPM1 mutations are associated with improved outcomes although the mechanism is not known. Concurrent mutations (such as an FTL3 mutation) may influence prognosis, but generally NPM1 patients without concurrent mutations have better prognoses (Schiffer, 2021).
The CCAAT/enhancer binding protein alpha (CEBPA) gene mutation is also common in AML. CEBPA encodes a transcription factor necessary for myeloid differentiation. This mutation is one of the two mutations associated with familial leukemia and consists of about 10% of AML cases. Familial AML with a CEBPA mutation has a phenotype similar to those of “sporadic AML with biallelic CEBPA mutations,” but most of the current data revolves around the assessment of CEBPA double mutations. CEBPA single mutations and hypermethylated CEBPA requires further study (Schiffer, 2021).
Isocitrate dehydrogenase (IDH) 1 and 2 mutations comprise approximately 15% of AML cases. These mutations are mutually exclusive with Tet Methylcytosine Dioxygenase 2 (TET2) and Wilms' tumor 1 (WT1) mutations but are commonly seen with NPM1 and DNA methyltransferase 3A (DNMT3A) mutations. Data on the prognoses of these mutations is varied (Schiffer, 2021).
KIT mutations also comprise about 6% of AML cases. KIT encodes the receptor for a stem cell factor, and prognoses are varied (Castells, 2021; Schiffer, 2021). Some researchers suggest that of all KIT mutations, the D816 mutation is the most unfavorable prognostic factor in AML patients (Yui et al., 2017).
Approximately eight percent of AML cases consist of WT1 gene mutations. WT1 encodes a transcriptional regulator for genes involved in maturation and growth. Again, the prognosis of this mutation is mixed (Schiffer, 2021), although some researchers strongly support the theory that WT1 mutations are associated with poor AML prognoses (Hou et al., 2010). The WT1 mutation status of AML patients may also change during disease progression. New research has suggested that after allogenic stem cell transplantation, AML relapse could be due to a gain in WT1 gene alterations and a “high mutation load” (Vosberg et al., 2018).
ASXL Transcriptional Regulator 1 (ASXL1) and ASXL Transcriptional Regulator 2 (ASXL2) may also be mutated in AML cases. ASXL1 has an unclear function, but it is speculated to be related to histone post-translational modifications. The frequency of ASXL1 is varied, as estimates range from 6% to 30%. Furthermore, ASXL1 mutations are mutually exclusive with NPM1 mutations, and ASXL2 mutations are associated with RUNX1 mutations (also known as AML1 or CBFA2) (Schiffer, 2021)
The DNMT3A gene amounts to 20%-22% of AML cases. This gene plays a role in epigenetic modifications for development and differentiation. Mutations in this gene affect hematopoietic stem cell differentiation. Prognoses of this gene mutation have been mixed (Schiffer, 2021).
Tumor protein 53 (TP53) and RAS and may also be present in AML cases and may be accompanied by other genetic abnormalities. RAS regulates cell signal transduction, and its mutation leads to a constitutionally active growth stimulus whereas TP53 encodes a transcriptional activator of growth inhibitory genes (Frucht & Lucas, 2022; Rai & Stilgenbauer, 2021).
Gene expression profiling and microRNA expression profiling may also contribute to assessment and management of AML. Gene expression profiling has been used to differentiate between risk groups based on cytogenetic evaluation whereas microRNA profiling evaluates the regulation of gene expression. However, neither technique is used regularly in clinical practice as these techniques have yet to be widely validated (Schiffer, 2021).
Analytic Validity
There is very limited published literature on the analytic validity and clinical validity of genetic testing for FLT3 and NPM1 mutations in AML. However, the analytic validity of PCR in general is extremely high (Leonard, 2016). Other tools, such as flow cytometry and next generation sequencing (NGS) have also been used for AML prognostic and diagnostic purposes.
Ampasavate et al. (2019) have developed a quantitative protocol and flow cytometry-based method for monitoring an anti-FLT3 interaction. The FLT3 biomarker has been previously identified as a poor prognostic marker for AML patients. This method can rapidly identify intact FLT3 on the leukemic cell surface. “The results demonstrated good linearity (r2 > 0.99)”; further, “when compared with Western blotting results, FLT3 protein expression levels in leukemia patient's bone marrow samples were demonstrated in the same trend” (Ampasavate et al., 2019). The researchers state that this technique is reliable, rapid, effective and “provided a practical analysis of FLT-3 biomarker levels which is valuable for physician decision in acute leukemia treatment” (Ampasavate et al., 2019).
Alonso et al. (2019) have researched the utility of a 19-gene NGS panel for AML diagnostic purposes. This targeted NGS panel was studied in a cohort of 162 patients with AML. The authors note that “The assay yielded a median read depth > 2000×, with 88% of on-target reads and a mean uniformity > 93% without significant global strand bias. The method was sensitive and specific, with a valid performance at the clinical variant allele frequency cutoff of 3% for point mutations and 5% for insertions or deletions.” The researchers conclude that this is a “reliable and reproducible method” for AML diagnoses (Alonso et al., 2019).
Clinical Utility and Validity
The clinical utility of testing fallows for further risk stratification, prognostication, and guide management decisions in patients with AML. Several studies have concluded that FLT3 and NPM1 mutation testing in cytogenetically normal AML is useful for prognosis and treatment decision making (DeZern et al., 2011; Pastore et al., 2014; Willemze et al., 2014).
Devillier et al. (2015) sought to identify biological and prognostic subgroups based on genetic mutations in AML patients. A total of 125 AML patients with myelodysplasia-related changes (“MRC”) were evaluated. The authors focused on the 26 patients with ASXL1 mutations and 28 with TP53 mutations. The ASXL1 mutation cohort was found to have a higher proportion of marrow dysgranulopoiesis and an overall survival (OS) rate that was below average for wild-types (14% for ASXL1 mutants, 37% for wild-types). The TP53 cohort was found to have a “complex karyotype” and predicted a poor outcome with unfavorable cytogenetic risk AML. Both mutations were found to be an independent factor associated with shorter OS (Devillier et al., 2015).
Bolouri et al. (2018) examined 993 children’s genetic data from the Children's Oncology Group (COG) AML trials to characterize the molecular landscape of AML. The authors found that certain somatic variants, such as MBNL1, were “disproportionately prevalent” in children compared to adults. However, certain variants common in adults such as TP53, were not found in children. Other mutations such as NRAS and KRAS were “frequent” in pediatric AML. The authors concluded that their results “highlight the need for and facilitate the development of age-tailored targeted therapies for the treatment of pediatric AML” (Bolouri et al., 2018).
Jongen-Lavrencic et al. (2018) conducted a study of 482 patients 18 to 65 years with newly diagnosed AML. Targeted next-generation sequencing (NGS) was carried out at diagnosis and after induction therapy (during complete remission). At least one mutation was detected in 430 (89.2%) patients, and mutations persisted in 51.4% of those patients during complete remission. The detection of minimal residual disease was associated with a significantly increased relapse rate than no detection. Persistent DTA mutations (mutations in DNMT3A, TET2, and ASXL1) were not correlated with an increased relapse rate. Overall, the authors concluded, “A comparison of sequencing with flow cytometry for the detection of residual disease showed that sequencing had significant additive prognostic value. Among patients with AML, the detection of molecular minimal residual disease during complete remission had significant independent prognostic value with respect to relapse and survival rates, but the detection of persistent mutations that are associated with clonal hematopoiesis did not have such prognostic value within a 4-year time frame” (Jongen-Lavrencic et al., 2018).
Kuwatsuka et al. (2018) evaluated the genetic background of 103 young adults and their subsequent clinical outcomes. The 103 cases included mutations in FLT3-ITD, KIT, CEBPA, NRAS, KRAS, WT1, MLL-PTD, and NPM1. Overall, FLT3-ITD and NPM1 mutations were associated with a greater mortality risk. NPM1 mutations conferred a 100% survival rate in the absence of FLT3-ITD mutations, but FLT3-ITD conferred only a 35% survival without NPM1 mutations (Kuwatsuka et al., 2018).
Zhu et al. (2017) assessed the effect of gene mutations on the subsequent cytogenetic aberrations. A total of 560 patients were enrolled, and the authors examined the following alterations: “CEBPA, NPM1, FLT3, C-KIT, NRAS, WT1, DNMT3A, MLL-PTD and IDH1/2, as well as expression levels of MECOM, ERG, GATA2, WT1, BAALC, MEIS1 and SPI1.” The investigators found that the expression levels of MECOM, MEIS1, and BAALC influenced cytogenetic aberration. Further, FLT3, C-KIT, and NRAS mutations all contained a “conversed” expression profile of MEIS1, WT1, GATA2, and BAALC expression. The investigators also noted “FLT3, DNMT3A, NPM1 and biallelic CEBPA represented the mutations associated with the prognosis of AML in our group” (Zhu et al., 2017).
Papaemmanuil et al. (2016) examined the relationship between genotype and pathophysiology in AML. A total of 1540 patients with 5234 driver mutations across 76 genes were studied. The authors found three genomic subcategories in addition to the currently defined AML subgroups: mutations in genes encoding chromatin, RNA splicing regulators or both (such as ASXL1 or RUNX1), TP53 mutations, chromosomal aneuploidies or both (unusual karyotypes and TP53), and IDH2 mutations. The authors noted that “patients with chromatin–spliceosome and TP53–aneuploidy AML had poor outcomes.” The NPM1 cohort was the largest of the sample (27%, 436 patients) and 319 of those patients also carried a DNA methylation or hydroxylmethylation gene, such as IDH1/2 or TET2. The authors also noted that NPM1-DNMT3A–NRASG12/13 had an “unexpectedly benign” prognosis, and the NPM1 subgroup’s prognoses were largely determined by the context in which the NPM1 mutation occurred (such as in NRAS, IDH, and so on) (Papaemmanuil et al., 2016).
Sperr et al. (2016) evaluated the effect of a genetic mutation and karyotype on the efficacy of treatment for elderly patients. A total of 192 patients over 60 years old were enrolled, and 115 of these patients achieved “complete hematologic remission (CR).” The authors stated that NPM1 mutations (NPM1mut) and karyotype were the only independent predictors of survival, also noting that NPM1mut showed a prognostic impact on both normal (CN) and non-chromosomal (Mkneg) karyotypes. The authors concluded that “elderly patients with CN/Mkneg-NPM1mut or core binding factor AML can achieve long term median continuous CR when treated with intensive induction and consolidation therapy whereas most elderly patients with CN/Mkneg-NPM1wild-type or CN/Mkpos AML may not benefit from intensive chemotherapy” (Sperr et al., 2016).
Heiblig et al. (2019) assessed the impact of NPM1 subtypes on treatment outcomes. One hundred seventy-five patients were examined. The authors found that out of the NPM1 AML cases, 73% (128) were “Type A” mutations (TCTG at exon 12) and 27% (47) were “Non Type-A mutations” (Type B: CATG and Type D: CGTG). The Type-A mutations were found to achieve minimal residual disease (MRD) earlier than non Type-A mutations. However, non-type A mutations achieved better rates of medial survival (Heiblig et al., 2019).
Xu et al. (2020) have analyzed data from 220 normal karyotype AML pediatric patients. Participants were selected from the Cancer Genome Atlas database. It was found that 12.7% of these patients had WT1 mutations, and that “the WT1-mutated group suffered lower rates of complete remission (CR) (P < 0.001 and P < 0.001, respectively) but higher rates of minimal residual disease (MRD) (P = 0.003 and P = 0.021, respectively) after both one and two courses of induction chemotherapy” (Xu et al., 2020). Patients with WT1 mutations also had significantly worse event-free and overall survival rates (P=0.007 and P<0.001, respectively) (Xu et al., 2020).
Sasaki et al. (2020) studied the impact of ASXL1, DNMT3A, JAK2, TET2, and TP53 mutations on survival in newly diagnosed acute myeloid leukemia (AML) patients. 421 bone marrow aspirates were studied using NGS for these mutations with a minimum variant allele frequency (VAF) of 5%. "A total of 71 patients (17%) had ASXL1 mutations, 104 patients (25%) had DNMT3A mutations, 16 patients (4%) had JAK2 mutations, 82 patients (20%) had TET2 mutations, and 86 patients (20%) had TP53 mutations." The median VAF of ASXL1 was 34.31% (range, 1.17% – 58.62%), DNMT3A was 41.76% (range, 1.02%‐91.66%), JAK2 was 46.70% (range, 10.4% – 71.7%), TET2 was 42.78% (range, 2.26%‐95.32%), and TP53 was 45.47% (range, 1.15%‐93.74%). In patients with these mutations, the median overall survival was 11 months. In patients without these mutations, the overall survival was 27 months. The authors conclude that "The VAF of ASXL1, DNMT3A, JAK2, TET2, TP53, and NPM1 mutations is associated with worse prognosis in patients with newly diagnosed AML." (Sasaki et al., 2020).
Duncavage et al. (2021) investigated the clinical utility and accuracy of whole-genome sequencing (WGS) for the purpose of risk stratification in patients with acute myeloid leukemia (AML) or myelodysplastic syndromes (MDS). The results from 263 patients were compared with findings from cytogenetic analysis and targeted sequencing. When conducting the WGS, they found that all 40 recurrent translocations and 91 copy-number alterations found on cytogenetic analysis were identified on WGS. There were also new clinically reportable genomic events among 17.0% of the patient sample. The standard AML risk groups, “as defined by sequencing results instead of cytogenetic analysis, correlated with clinical outcomes.” WGS was also able to classify patients with inconclusive cytogenetic analysis results into risk groups. This demonstrates that WGS could increase the diagnostic yield of AML and MDS and supplement the accuracy of cytogenetic analysis (Duncavage et al., 2021).
World Health Organization (WHO)
In the recent revision of the 4th edition on Classification of Tumors of Hematopoietic and Lymphoid Tissues, World Health Organization (WHO) incorporated new molecular genetic findings and clinical data into its classification of acute leukemias. WHO expanded on the prognostic significance of various gene mutations for each AML subtype. For example, for the AML with recurrent genetic abnormalities, inv(3) (q21.3; q26.2) does not represent a fusion gene, but rather a repositioning of a distal GATA2 enhancer leading to activation of MECOM (EVI1) expression and GATA2 haploinsufficiency. The AML with CEBPA mutation is defined based on biallelic mutation instead of single mutations because of prognostic significance. The provisional two categories are also added such as AML with RUNX1 for de novo AML without preexisting cytogenetic abnormalities associated with MDS and AML with BCR-ABL1 fusion gene. AML with NPM1 or biallelic CEBPA mutations and multilineage dysplasia are now considered separately instead of being a part of AML with myelodysplasia-related changes because of a lack of prognostic significance. The complete list for acute myeloid neoplasms 2016 WHO classification is shown in Figure 1 (Arber et al., 2016).
In the absence of JAK2, CALR, and MPL mutations, the presence of another clonal marker is included as one of the major diagnostic criteria for PMF. Additional mutation in ASXL1, EZH2, TET2, IDH1, IDH2, SRSF2 and SF3B1 genes are noted to be of use in determining the clonal nature of the disease.
Figure 1: WHO classification of AML and related neoplasms from Arber et al. (2016)

College of American Pathologists (CAP) and American Society of Hematology (ASH)
Following recent progress in molecular genetic findings and 2016 WHO classification of acute leukemias, the College of American Pathologists (CAP) and the American Society of Hematology (ASH) have formed an expert panel to review and establish guidelines for appropriate laboratory testing. The published guideline provides twenty-seven guideline statements ranging from recommendations on what testing is appropriate for the diagnostic and prognostic evaluation of leukemias to where the testing should be performed and how results should be reported. The appropriate molecular genetic testing for AML is discussed starting from 16th guideline statement.
Expert panel strongly recommends testing for FLT3-ITD in adult and pediatric patients with suspected or confirmed AML of any type. They also recommend testing for other mutational analysis that could include, but not limited to, IDH1, IDH2, TET2, WT1, DNMT3A, and/or TP53 for prognostic and/or therapeutic purposes (Statement 16).
In the 17th guideline statement, expert panel strongly recommends testing for KIT mutation in adult patients with confirmed core-binding factor (CBF) AML (AML with t(8;21)(q22;q22.1); RUNX1-RUNX1T1 or inv(16)(p13.1q22) /t(16;16)(p13.1;q22); CBFB-MYH11). It is only an expert consensus opinion for testing KIT mutation in pediatric patients with confirmed core binding factor AML (AML with t(8;21)(q22;q22.1); RUNX1- RUNX1T1 or inv(16)(p13.1q22) /t(16;16)(p13.1;q22); CBFB-MYH11) which is not a strong recommendation (Statement 17).
The strong recommendation is also given for patients other than those with confirmed core binding factor AML, APL, or AML with myelodysplasia-related cytogenetic abnormalities that testing is needed for mutational analysis for NPM1, CEBPA, and RUNX1 (Statement 19).
In the 20th guideline statement, expert panel is providing no recommendation on either for or against the use of global/gene specific methylation, microRNA (miRNA) expression, or gene expression analysis for diagnosis or prognosis in patients with confirmed acute leukemia.
Finally, in their last statement, expert panel strongly recommends the use of current WHO terminology for the final diagnosis and classification of acute leukemias (Arber et al., 2017).
American Society of Clinical Oncology (ASCO)
The ASCO has announced an endorsement of the 2017 CAP and ASH guideline regarding the initial diagnostic work-up of acute leukemia. The ASCO supports all twenty-seven guideline statements. The statements relevant to this policy are noted in the 2017 CAP and ASH guidelines above.
National Comprehensive Cancer Network (NCCN)
For the initial evaluation of AML, the NCCN guidelines recommend bone marrow core biopsy and aspirate analyses “(including immunophenotyping by immunohistochemistry stains with flow cytometry)” and cytogenetic analyses (karyotype + FISH), molecular analyses (ASXL1, c-KIT, FLT3 [ITD and TKD], NPM1, CEBPA (biallelic), IDH1, IDH2, RUNX1, TP53 and other mutations) (NCCN, 2021).
The NCCN also states that “Several gene mutations are associated with specific prognoses in a subset of patients (category 2A) and may guide treatment decisions (category 2B). Presently, c-KIT, FLT3-ITD, FLT3-TKD, NPM1, CEBPA (biallelic), IDH1/IDH2, RUNX1, ASXL1, TP53, BCR-ABL, and PML-RAR alpha are included in this group” (NCCN, 2021).
European LeukemiaNet (ELN) Working Party
The ELN expert panel released guidelines for the assessment of measurable residual disease (MRD) of AML that were updated in 2021. First, for molecular MRD recommendations, “techniques for molecular MRD assessment should reach an LOD [limit of detection] of 10-3 or lower. To achieve this LOD, qPCR, dPCR, or error-corrected NGS using UMIs [unique molecular identifiers] is recommended (Grade of Recommendation: B).”
Other recommendations for molecular MRD testing included:
- “For patients with mutant NPM1, CBF AML (RUNX1-RUNX1T1 or CBFB-MYH11), or APL (PML-RARA), we recommend molecular MRD assessment by qPCR or dPCR. (Grade of Recommendation: A)”
- “Leukemia-specific PCR assays (e.g., for NPM1, PML-RARA, or CBF AML) are preferred over fewer specific markers, such as WT1 or EV11 expression (Grade of Recommendation: B)”
- “Targeted NGS-MRD using specific mutations identified at diagnosis vs agnostic panel approaches has different strengths and limitations, but both approaches can be considered, depending on sensitivity, turnaround time, resource use, setting (research, clinical trial, or clinical routine), and ability to standardize methodology and reporting. (Grade of Recommendation: B)”
- “If a panel approach is used for NGS-MRD, emerging variants not found at diagnosis should be reported only if confidently detected above background noise. (Grade of Recommendation: B)”
- “For NGS-MRD, we recommend considering all detected mutations as potential MRD markers, with the limitations detailed in recommendations B9 to B11 (listed below) (Grade of Recommendation: B)”
- “Germline mutations (VAF of ∼ 50 in genes ANKRD26, CEBPA, DDX41, ETV6, GATA2, RUNX1, and TP53) should be excluded as NGS-MRD markers, as they are noninformative for MRD. (Grade of Recommendation: A)”
- “Mutations in DNMT3A, TET2, and ASXL1 (DTA) can be found in age-related clonal hematopoiesis and should be excluded from MRD analysis. (Grade of Recommendation: A)”
- “Mutations in signaling pathway genes (FLT3-ITD, FLT3-TKD, KIT, and RAS, among others) most likely represent residual AML when detected, but are often subclonal and have a low negative predictive value. These mutations are best used in combination with additional MRD markers. (Grade of Recommendation: B)” (Heuser et al., 2021)
The ELN also released guidelines for the diagnosis and management of AML. Their genetic test battery includes cytogenetics, screening for gene mutations in NPM1, CEBPA, RUNX1, FLT3, TP53, ASXL1, and screening for gene rearrangements in PML-RARA, CBFB-MYH11, RUNX1-RUNX1T1, BCR-ABL1, other fusion genes (if available) (Döhner et al., 2017).
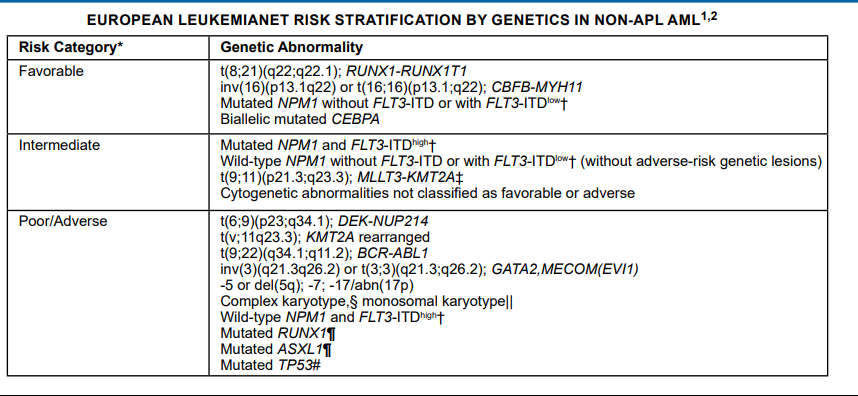
British Committee for Standards in Haematology (BCSH)
The BCSH has published guidelines for the diagnosis and management of AML in pregnancy. These guidelines state that “The diagnostic criteria for AML are the same in a pregnant patient as in non-pregnant women. These criteria are defined in the World Health Organization (WHO) classification of the myeloid neoplasms. Where a diagnosis of leukemia is suspected, care must be taken to ensure that marrow samples are directed for immunophenotypic, cytogenetic and molecular analysis to allow accurate sub-typing and understanding of prognostic features” (Ali et al., 2015).
Canadian Consensus Guidelines
Revised Canadian consensus guidelines for the treatment of older patients with AML were published in 2017. The guidelines included the following recommendation:
- “For older patients who are candidates for intensive chemotherapy, FLT-ITD and TKD mutation testing results should be provided within one week. For patients up to age 70 with a FLT3-ITD or TKD mutation, midostaurin, if available, should be added to induction and consolidation, and continued as maintenance therapy if not transplanted, in the schedule used in the RATIFY and German AMLSG studies” (Brandwein et al., 2017).
European Society For Medical Oncology (ESMO)
ESMO released guidelines for the diagnosis of acute myeloid leukemia in adult patients. They recommend molecular testing for c-KIT, FLT3-ITD, FLT3-TKD, NPM1, CEBPA, IDH1, IDH2, and TP53 mutations. They state that “next-generation sequencing (NGS) of a panel of genes commonly mutated in AML provides important additional prognostic and therapeutic information” (Heuser et al., 2020).
Table of Terminology
|
Term |
Definition |
|
AML |
Acute myeloid leukemia |
|
ANKRD26 |
Ankyrin repeat domain containing 26 |
|
APL |
Acute promyelocytic leukemia |
|
ASCO |
American Society of Clinical Oncology |
|
ASH |
American Society of Hematology |
|
ASXL1 |
ASXL transcriptional regulator 1 |
|
ASXL2 |
ASXL transcriptional regulator 2 |
|
BAALC |
Brain and acute leukemia cytoplasmic |
|
BCR-ABL1 |
BCR activator of RhoGEF and GTPase- ABL proto-oncogene 1, non-receptor tyrosine kinase |
|
BCSH |
British Committee for Standards in Haematology |
|
c-KIT |
Commonly used alias for KIT proto-oncogene, receptor tyrosine kinase |
|
CAP |
College of American Pathologists |
|
CBF |
Core-binding factor |
|
CALR |
Calreticulin |
|
CBFA2 |
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
|
CEBPA |
CCAAT/enhancer binding protein alpha |
|
CLIA ’88 |
Clinical Laboratory Improvement Amendments of 1988 |
|
CMS |
Centers for Medicare and Medicaid |
|
CN |
Normal karyotypes |
|
CR |
Complete hematologic remission |
|
DDX41 |
DEAD-box helicase 41 |
|
DNA |
Deoxyribonucleic acid |
|
DNMT3A |
Deoxyribonucleic acid methyltransferase 3A |
|
ELN |
European LeukemiaNet |
|
EMSO |
European Society For Medical Oncology |
|
ERG |
ETS transcription factor ERG |
|
EVI1 |
Ecotropic virus integration site 1 protein homolog |
|
EZH2 |
Enhancer of zeste 2 polycomb repressive complex 2 subunit |
|
FDA |
Food and Drug Administration |
|
FLT3 |
FMS-like tyrosine kinase 3 |
|
GATA2 |
GATA binding protein 2 |
|
IDH1 |
Isocitrate dehydrogenase 1 |
|
IDH2 |
Isocitrate dehydrogenase 2 |
|
ITD |
Internal tandem duplications |
|
JAK2 |
Janus kinase 2 |
|
KIT |
KIT proto-oncogene, receptor tyrosine kinase |
|
KRAS |
KRAS proto-oncogene, GTPase |
|
LTD |
Laboratory-developed test |
|
MBNL1 |
Muscleblind like splicing regulator 1 |
|
MECOM |
MDS1 and EVI1 complex locus |
|
MEIS1 |
Muscleblind like splicing regulator 1 |
|
miRNA |
Micro ribonucleic acid |
|
Mkneg |
Non-chromosomal karyotype |
|
MLL-PTD |
Mll -Partial Tandem Duplication |
|
MPL |
MPL proto-oncogene, thrombopoietin receptor |
|
MRD |
Minimal residual disease |
|
MYH11 |
Myosin heavy chain 11 |
|
NCCN |
National Comprehensive Cancer Network |
|
NGS |
Next generation sequencing |
|
NPM1 |
Nucleophosmin 1 |
|
NPM1mut |
NPM1 mutations |
|
NRAS |
NRAS proto-oncogene, GTPase |
|
PCR |
Polymerase chain reaction |
|
PMF |
Polyamine modulated factor 1 |
|
PML-RAR |
Promyelocytic leukemia-retinoic acid receptor |
|
RNA |
Ribonucleic acid |
|
RT-PCR |
Reverse transcription- polymerase chain reaction |
|
RUNX1 |
RUNX family transcription factor 1 |
|
RUNX1T1 |
RUNX1 partner transcriptional co-repressor 1 |
|
SPI1 |
Spi-1 proto-oncogene |
|
SF3B1 |
Splicing factor 3b subunit 1 |
|
SRSF2 |
Serine and arginine rich splicing factor 2 |
|
TET2 |
Tet methylcytosine dioxygenase 2 |
|
TKD |
Tyrosine kinase domain |
|
TP53 |
Tumor protein 53 |
|
WHO |
World Health Organization |
|
WT1 |
Wilms' tumor 1 |
References
- Ali, S., Jones, G. L., Culligan, D. J., Marsden, P. J., Russell, N., Embleton, N. D., & Craddock, C. (2015). Guidelines for the diagnosis and management of acute myeloid leukaemia in pregnancy. Br J Haematol, 170(4), 487-495. https://doi.org/10.1111/bjh.13554
- Alonso, C. M., Llop, M., Sargas, C., Pedrola, L., Panadero, J., Hervás, D., Cervera, J., Such, E., Ibáñez, M., Ayala, R., Martínez-López, J., Onecha, E., de Juan, I., Palanca, S., Martínez-Cuadrón, D., Rodríguez-Veiga, R., Boluda, B., Montesinos, P., Sanz, G., . . . Barragán, E. (2019). Clinical Utility of a Next-Generation Sequencing Panel for Acute Myeloid Leukemia Diagnostics. J Mol Diagn, 21(2), 228-240. https://doi.org/10.1016/j.jmoldx.2018.09.009
- Ampasavate, C., Jutapakdee, W., Phongpradist, R., Tima, S., Tantiworawit, A., Charoenkwan, P., Chinwong, D., & Anuchapreeda, S. (2019). FLT3, a prognostic biomarker for acute myeloid leukemia (AML): Quantitative monitoring with a simple anti-FLT3 interaction and flow cytometric method. J Clin Lab Anal, 33(4), e22859. https://doi.org/10.1002/jcla.22859
- Arber, D. A., Borowitz, M. J., Cessna, M., Etzell, J., Foucar, K., Hasserjian, R. P., Rizzo, J. D., Theil, K., Wang, S. A., Smith, A. T., Rumble, R. B., Thomas, N. E., & Vardiman, J. W. (2017). Initial Diagnostic Workup of Acute Leukemia: Guideline From the College of American Pathologists and the American Society of Hematology. Arch Pathol Lab Med, 141(10), 1342-1393. https://doi.org/10.5858/arpa.2016-0504-CP
- Arber, D. A., Orazi, A., Hasserjian, R., Thiele, J., Borowitz, M. J., Le Beau, M. M., Bloomfield, C. D., Cazzola, M., & Vardiman, J. W. (2016). The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood, 127(20), 2391-2405. https://doi.org/10.1182/blood-2016-03-643544
- Bolouri, H., Farrar, J. E., Triche, T., Jr., Ries, R. E., Lim, E. L., Alonzo, T. A., Ma, Y., Moore, R., Mungall, A. J., Marra, M. A., Zhang, J., Ma, X., Liu, Y., Liu, Y., Auvil, J. M. G., Davidsen, T. M., Gesuwan, P., Hermida, L. C., Salhia, B., . . . Meshinchi, S. (2018). The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med, 24(1), 103-112. https://doi.org/10.1038/nm.4439
- Brandwein, J. M., Zhu, N., Kumar, R., Leber, B., Sabloff, M., Sandhu, I., Kassis, J., Olney, H. J., Elemary, M., & Schuh, A. C. (2017). Treatment of older patients with acute myeloid leukemia (AML): revised Canadian consensus guidelines. Am J Blood Res, 7(4), 30-40. http://www.ajblood.us/files/ajbr0058846.pdf
- Castells, M. (2021, 07/15/2021). Mastocytosis (cutaneous and systemic) in adults: Epidemiology, pathogenesis, clinical manifestations, and diagnosis. https://www.uptodate.com/contents/mastocytosis-cutaneous-and-systemic-in-adults-epidemiology-pathogenesis-clinical-manifestations-and-diagnosis
- CGARN. (2013). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med, 368(22), 2059-2074. https://doi.org/10.1056/NEJMoa1301689
- Devillier, R., Mansat-De Mas, V., Gelsi-Boyer, V., Demur, C., Murati, A., Corre, J., Prebet, T., Bertoli, S., Brecqueville, M., Arnoulet, C., Recher, C., Vey, N., Mozziconacci, M. J., Delabesse, E., & Birnbaum, D. (2015). Role of ASXL1 and TP53 mutations in the molecular classification and prognosis of acute myeloid leukemias with myelodysplasia-related changes. Oncotarget, 6(10), 8388-8396. https://doi.org/10.18632/oncotarget.3460
- DeZern, A. E., Sung, A., Kim, S., Smith, B. D., Karp, J. E., Gore, S. D., Jones, R. J., Fuchs, E., Luznik, L., McDevitt, M., & Levis, M. (2011). Role of allogeneic transplantation for FLT3/ITD acute myeloid leukemia: outcomes from 133 consecutive newly diagnosed patients from a single institution. Biol Blood Marrow Transplant, 17(9), 1404-1409. https://doi.org/10.1016/j.bbmt.2011.02.003
- Döhner, H., Estey, E., Grimwade, D., Amadori, S., Appelbaum, F. R., Büchner, T., Dombret, H., Ebert, B. L., Fenaux, P., Larson, R. A., Levine, R. L., Lo-Coco, F., Naoe, T., Niederwieser, D., Ossenkoppele, G. J., Sanz, M., Sierra, J., Tallman, M. S., Tien, H.-F., . . . Bloomfield, C. D. (2017). Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood, 129(4), 424. https://doi.org/10.1182/blood-2016-08-733196
- Duncavage, E. J., Schroeder, M. C., O'Laughlin, M., Wilson, R., MacMillan, S., Bohannon, A., Kruchowski, S., Garza, J., Du, F., Hughes, A. E. O., Robinson, J., Hughes, E., Heath, S. E., Baty, J. D., Neidich, J., Christopher, M. J., Jacoby, M. A., Uy, G. L., Fulton, R. S., . . . Spencer, D. H. (2021). Genome Sequencing as an Alternative to Cytogenetic Analysis in Myeloid Cancers. N Engl J Med, 384(10), 924-935. https://doi.org/10.1056/NEJMoa2024534
- Falini, B., Mecucci, C., Tiacci, E., Alcalay, M., Rosati, R., Pasqualucci, L., La Starza, R., Diverio, D., Colombo, E., Santucci, A., Bigerna, B., Pacini, R., Pucciarini, A., Liso, A., Vignetti, M., Fazi, P., Meani, N., Pettirossi, V., Saglio, G., . . . Martelli, M. F. (2005). Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med, 352(3), 254-266. https://doi.org/10.1056/NEJMoa041974
- FDA. (2017a). Premarket Approval (PMA) for LeukoStrat CDx FLT3 Mutation Assay. U.S. Food and Drug Administration Retrieved from https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P160040
- FDA. (2017b). Premarket approval order for Abbott RealTime IDH2. U.S. Food and Drug Administration Retrieved from https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P170005
- Frucht, H., & Lucas, A. L. (2022, 01/21/2022). Molecular genetics of colorectal cancer. https://www.uptodate.com/contents/molecular-genetics-of-colorectal-cancer
- Heiblig, M., Sujobert, P., Hayette, S., Balsat, M., Elhamri, M., Salles, G., & Thomas, X. (2019). Impact of NPM1 mutation subtypes on treatment outcome in AML: The Lyon-University Hospital experience. Leuk Res, 76, 29-32. https://doi.org/10.1016/j.leukres.2018.11.016
- Heuser, M., Freeman, S. D., Ossenkoppele, G. J., Buccisano, F., Hourigan, C. S., Ngai, L. L., Tettero, J. M., Bachas, C., Baer, C., Béné, M.-C., Bücklein, V., Czyz, A., Denys, B., Dillon, R., Feuring-Buske, M., Guzman, M. L., Haferlach, T., Han, L., Herzig, J. K., . . . Cloos, J. (2021). 2021 Update on MRD in acute myeloid leukemia: a consensus document from the European LeukemiaNet MRD Working Party. Blood, 138(26), 2753-2767. https://doi.org/10.1182/blood.2021013626
- Heuser, M., Ofran, Y., Boissel, N., Brunet Mauri, S., Craddock, C., Janssen, J., Wierzbowska, A., & Buske, C. (2020). Acute myeloid leukaemia in adult patients: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol, 31(6), 697-712. https://doi.org/10.1016/j.annonc.2020.02.018
- Hou, H. A., Huang, T. C., Lin, L. I., Liu, C. Y., Chen, C. Y., Chou, W. C., Tang, J. L., Tseng, M. H., Huang, C. F., Chiang, Y. C., Lee, F. Y., Liu, M. C., Yao, M., Huang, S. Y., Ko, B. S., Hsu, S. C., Wu, S. J., Tsay, W., Chen, Y. C., & Tien, H. F. (2010). WT1 mutation in 470 adult patients with acute myeloid leukemia: stability during disease evolution and implication of its incorporation into a survival scoring system. Blood, 115(25), 5222-5231. https://doi.org/10.1182/blood-2009-12-259390
- Jongen-Lavrencic, M., Grob, T., Hanekamp, D., Kavelaars, F. G., Al Hinai, A., Zeilemaker, A., Erpelinck-Verschueren, C. A. J., Gradowska, P. L., Meijer, R., Cloos, J., Biemond, B. J., Graux, C., van Marwijk Kooy, M., Manz, M. G., Pabst, T., Passweg, J. R., Havelange, V., Ossenkoppele, G. J., Sanders, M. A., . . . Valk, P. J. M. (2018). Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N Engl J Med, 378(13), 1189-1199. https://doi.org/10.1056/NEJMoa1716863
- Kuwatsuka, Y., Tomizawa, D., Kihara, R., Nagata, Y., Shiba, N., Iijima-Yamashita, Y., Shimada, A., Deguchi, T., Miyachi, H., Tawa, A., Taga, T., Kinoshita, A., Nakayama, H., Kiyokawa, N., Saito, A. M., Koh, K., Goto, H., Kosaka, Y., Asou, N., . . . Kiyoi, H. (2018). Prognostic value of genetic mutations in adolescent and young adults with acute myeloid leukemia. Int J Hematol, 107(2), 201-210. https://doi.org/10.1007/s12185-017-2340-z
- Leonard, D. G. B. (2016). Molecular Pathology in Clinical Practice (D. G. B. Leonard, Ed.). Springer. http://www.springer.com/us/book/9783319196732
- NCCN. (2021, 12/02/2021). NCCN Clinical Practice Guidelines in Oncology; Acute Myeloid Leukemia Version 1.2022. https://www.nccn.org/professionals/physician_gls/pdf/aml.pdf
- Papaemmanuil, E., Gerstung, M., Bullinger, L., Gaidzik, V. I., Paschka, P., Roberts, N. D., Potter, N. E., Heuser, M., Thol, F., Bolli, N., Gundem, G., Van Loo, P., Martincorena, I., Ganly, P., Mudie, L., McLaren, S., O'Meara, S., Raine, K., Jones, D. R., . . . Campbell, P. J. (2016). Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med, 374(23), 2209-2221. https://doi.org/10.1056/NEJMoa1516192
- Pastore, F., Greif, P. A., Schneider, S., Ksienzyk, B., Mellert, G., Zellmeier, E., Braess, J., Sauerland, C. M., Heinecke, A., Krug, U., Berdel, W. E., Buechner, T., Woermann, B., Hiddemann, W., & Spiekermann, K. (2014). The NPM1 mutation type has no impact on survival in cytogenetically normal AML. PLoS One, 9(10), e109759. https://doi.org/10.1371/journal.pone.0109759
- Rai, K., & Stilgenbauer, S. (2021, 07/12/2021). Pathophysiology and genetic features of chronic lymphocytic leukemia. https://www.uptodate.com/contents/pathobiology-of-chronic-lymphocytic-leukemia
- Schiffer, C. (2021, 10/29/2021). Prognosis of acute myeloid leukemia. https://www.uptodate.com/contents/prognosis-of-acute-myeloid-leukemia
- Schiffer, C., & Gurbaxani, S. (2022, 12/21/2022). Clinical manifestations, pathologic features, and diagnosis of acute myeloid leukemia. https://www.uptodate.com/contents/clinical-manifestations-pathologic-features-and-diagnosis-of-acute-myeloid-leukemia
- Siegel, R. L., Miller, K. D., & Jemal, A. (2017). Cancer Statistics, 2017. CA Cancer J Clin, 67(1), 7-30. https://doi.org/10.3322/caac.21387
- Sperr, W. R., Zach, O., Poll, I., Herndlhofer, S., Knoebl, P., Weltermann, A., Streubel, B., Jaeger, U., Kundi, M., & Valent, P. (2016). Karyotype plus NPM1 mutation status defines a group of elderly patients with AML (>/=60 years) who benefit from intensive post-induction consolidation therapy. Am J Hematol, 91(12), 1239-1245. https://doi.org/10.1002/ajh.24560
- Stock, W. T., Michael. (2020). Molecular genetics of acute myeloid leukemia. In R. Larson (Ed.), UpToDate. https://www.uptodate.com/contents/molecular-genetics-of-acute-myeloid-leukemia?search=acute%20myeloid%20leukemia%20genetics&source=search_result&selectedTitle=1~150&usage_type=default&display_rank=1
- Vosberg, S., Hartmann, L., Metzeler, K. H., Konstandin, N. P., Schneider, S., Varadharajan, A., Hauser, A., Krebs, S., Blum, H., Bohlander, S. K., Hiddemann, W., Tischer, J., Spiekermann, K., & Greif, P. A. (2018). Relapse of acute myeloid leukemia after allogeneic stem cell transplantation is associated with gain of WT1 alterations and high mutation load. Haematologica, 103(12), e581-e584. https://doi.org/10.3324/haematol.2018.193102
- Willemze, R., Suciu, S., Meloni, G., Labar, B., Marie, J. P., Halkes, C. J., Muus, P., Mistrik, M., Amadori, S., Specchia, G., Fabbiano, F., Nobile, F., Sborgia, M., Camera, A., Selleslag, D. L., Lefrere, F., Sr., Magro, D., Sica, S., Cantore, N., . . . de Witte, T. (2014). High-dose cytarabine in induction treatment improves the outcome of adult patients younger than age 46 years with acute myeloid leukemia: results of the EORTC-GIMEMA AML-12 trial. J Clin Oncol, 32(3), 219-228. https://doi.org/10.1200/jco.2013.51.8571
- Xu, J., Zhang, Y., Hu, J., Ren, Y., & Wang, H. (2020). Clinical features and prognosis of normal karyotype acute myeloid leukemia pediatric patients with WT1 mutations: an analysis based on TCGA database. Hematology, 25(1), 79-84. https://doi.org/10.1080/16078454.2020.1720102
- Yamamoto, J. F., & Goodman, M. T. (2008). Patterns of leukemia incidence in the United States by subtype and demographic characteristics, 1997-2002. Cancer Causes Control, 19(4), 379-390. https://doi.org/10.1007/s10552-007-9097-2
- Yui, S., Kurosawa, S., Yamaguchi, H., Kanamori, H., Ueki, T., Uoshima, N., Mizuno, I., Shono, K., Usuki, K., Chiba, S., Nakamura, Y., Yanada, M., Kanda, J., Tajika, K., Gomi, S., Fukunaga, K., Wakita, S., Ryotokuji, T., Fukuda, T., & Inokuchi, K. (2017). D816 mutation of the KIT gene in core binding factor acute myeloid leukemia is associated with poorer prognosis than other KIT gene mutations. Ann Hematol, 96(10), 1641-1652. https://doi.org/10.1007/s00277-017-3074-y
- Zhu, Y. M., Wang, P. P., Huang, J. Y., Chen, Y. S., Chen, B., Dai, Y. J., Yan, H., Hu, Y., Cheng, W. Y., Ma, T. T., Chen, S. J., & Shen, Y. (2017). Gene mutational pattern and expression level in 560 acute myeloid leukemia patients and their clinical relevance. J Transl Med, 15(1), 178. https://doi.org/10.1186/s12967-017-1279-4
Coding Section
|
Code |
Number |
Code Description |
|
CPT |
81120 |
IDH1 (isocitrate dehydrogenase 1 [NADP+], soluble) (e.g., glioma), common variants (eg, R132H, R132C) |
|
|
81121 |
IDH2 (isocitrate dehydrogenase 2 [NADP+], mitochondrial) (e.g., glioma), common variants (eg, R140W, R172M) |
|
|
81175 |
ASXL1 (additional sex combs like 1, transcriptional regulator) (e.g., myelodysplastic syndrome, myeloproliferative neoplasms, chronic myelomonocytic leukemia), gene analysis; full gene sequence |
|
|
81176 |
ASXL1 (additional sex combs like 1, transcriptional regulator) (e.g., myelodysplastic syndrome, myeloproliferative neoplasms, chronic myelomonocytic leukemia), gene analysis; targeted sequence analysis (e.g., exon 12) |
|
|
81218 |
CEBPA (CCAAT/enhancer binding protein [C/EBP], alpha) (e.g., acute myeloid leukemia), gene analysis, full gene sequence |
|
|
81245 |
FLT3 (fms-related tyrosine kinase 3) (e.g., acute myeloid leukemia), gene analysis; internal tandem duplication (ITD) variants (i.e., exons 14, 15) |
|
|
81246 |
FLT3 (fms-related tyrosine kinase 3) (e.g., acute myeloid leukemia), gene analysis; tyrosine kinase domain (TKD) variants (e.g., D835, I836) |
|
|
81272 |
KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog) (eg, gastrointestinal stromal tumor [GIST], acute myeloid leukemia, melanoma), gene analysis, targeted sequence analysis (e.g., exons 8, 11, 13, 17, 18) |
|
|
81273 |
KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog) (e.g., mastocytosis), gene analysis, D816 variant(s) |
|
|
81310 |
NPM1 (nucleophosmin) (e.g., acute myeloid leukemia) gene analysis, exon 12 variants |
|
|
81334 |
RUNX1 (runt related transcription factor 1) (e.g., acute myeloid leukemia, familial platelet disorder with associated myeloid malignancy), gene analysis, targeted sequence analysis (e.g., exons 3-8) |
|
|
81403 |
Molecular pathology procedure, Level 4 Gene: |
|
|
81404 |
Molecular pathology procedure, Level 5 Gene: |
|
|
81405 |
Molecular pathology procedure, Level 6 Gene: |
|
|
81479 |
Unlisted molecular pathology procedure Gene: |
|
|
0023U |
Oncology (acute myelogenous leukemia), DNA, genotyping of internal tandem duplication, p.D835, p.I836, using mononuclear cells, reported as detection or non-detection of FLT3 mutation and indication for or against the use of midostaurin |
|
|
0046U |
FLT3 (fms-related tyrosine kinase 3) (e.g., acute myeloid leukemia) internal tandem duplication (ITD) variants, quantitative |
|
|
0049U |
NPM1 (nucleophosmin) (e.g., acute myeloid leukemia) gene analysis, quantitative |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community, Blue Cross Blue Shield Association technology assessment program (TEC) and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies, and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2014 Forward
| 08/01/2022 | Annual review, no change to policy intent. Updating coding, description, rationale and references. |
|
07/27/2021 |
Annual review, removing Genetic testing for FLT3, NPM1, CEBPA, IDH 1/2, KIT and other mutations to detect minimal residual disease is investigational/unproven therefore considered NOT MEDICALLY NECESSARY. as this is addressed in another policy. No other change to policy intent. Updating rationale and references. Removing regulatory status as that is included in the rationale. |
|
07/15/2020 |
Annual review, updating policy for clarity; also updating background, rationale and references. |
|
07/12/2019 |
Annual review, no change to policy intent. Updating guidelines. |
|
07/18/2018 |
Annual review, adding variant ASXL1 as medically necessary, updating coding section. No other changes made. |
|
12/7/207 |
Updating policy with 2018 coding. No other changes. |
|
12/05/2017 |
Interim review. Updating policy verbiage and adding new codes.. |
|
07/18/2017 |
Annual review, updating title and coding. Adding medical necessity criteria for NPM1, CEPBA, IDH 1/2 mutations and KIT mutations. Updating investigational policy status. |
|
04/25/2017 |
Updated category to Laboratory. No other changes. |
|
12/05/2016 |
Annual review, no change to policy intent. |
|
12/1/2015 |
Updated CPT codes for 2016. |
|
11/12/2015 |
Annual review, no change to policy intent. Updated background, description, guidelines, rationale, references and coding. Added appendix 1. |
|
12/02/2014 |
New Policy |