Genetic Testing for Lactase Insufficiency - CAM 300
Description
Lactose intolerance (LI) is a common clinical syndrome defined by abdominal pain, flatulence, bloating, borborygmus and osmotic diarrhea; LI is caused by the breakdown of nondigested lactose by the gut microflora (Ponte et al., 2016a).
Lactose malabsorption (LM) is the non-digestion of lactose caused by low expression of the enzyme lactase and is a physiologic feature occurring in most mammals after infancy (Di Rienzo et al., 2013; Ponte et al., 2016b).
Policy
- The use of targeted mutation analysis of -13910 C>T for the prediction of lactase insufficiency is investigational/unproven and therefore considered NOT MEDICALLY NECESSARY.
- Genetic testing of the LCT gene and/or MCM6 gene for lactose intolerance and/or lactase insufficiency is investigational/unproven and therefore considered NOT MEDICALLY NECESSARY.
Table of Terminology
|
Term |
Definition |
|
AAP |
American Academy of Pediatrics |
|
ACG |
American College of Gastroenterology |
|
BSACI |
British Society for Allergy & Clinical Immunology |
|
C/T-13910 |
Eurasian lactase persistence variant |
|
CLIA |
Clinical Laboratory Improvement Amendments |
|
CMS |
Centers for Medicare and Medicaid Services |
|
FODMAP |
Fermentable oligo-, di- and monosaccharides and polyols |
|
FGID |
Functional gastrointestinal disorders |
|
HBT |
Hydrogen breath test |
|
IBD |
Inflammatory bowel disease |
|
LBT |
Lactose breath test |
|
LCT |
Lactase /lactase-phlorizin hydrolase |
|
LD |
Lactase deficiency |
|
LDTs |
Laboratory developed tests |
|
LI |
Lactose intolerance |
|
LIP |
Lactose-intolerant phenotype |
|
LM |
Lactose malabsorption |
|
LNP |
Lactase non-persistence |
|
MCM6 |
Minichromosome maintenance complex component 6 |
|
NIH |
National Institutes of Health |
|
PCR |
Polymerase chain reaction |
|
RNA |
Ribonucleic acid |
|
RT-PCR |
Real-time polymerase chain reaction |
|
SNPs |
Single nucleotide polymorphisms |
Rationale
Lactose is a disaccharide, which is a class of sugars comprised of two monosaccharides or simple sugars. Lactose consists of galactose bound to glucose and is the main source of carbohydrates from mammalian milk. Intestinal absorption of lactose requires initial hydrolysis by the enzyme lactase. Low lactase activity results in undigested lactose and colonic bacterial fermentation of that lactose. This leads to the characteristic symptoms of lactose intolerance, such as bloating and flatulence (Luyt et al., 2014).
Lactase expression decreases as consequence of the normal maturational down-regulation after weaning, ultimately to undetectable levels in most populations (Swallow, 2003). Lactase expression persists, however, in descendants of populations that traditionally practice cattle domestication who maintain the ability to digest milk and other dairy products into adulthood (Deng, Misselwitz, Dai, & Fox, 2015). Adult expression of the gene encoding lactase (LCT), located on chromosome two at 2q21 appears to be regulated by cis-acting elements (Wang et al., 1995) and is inherited as an autosomal recessive trait (Enattah et al., 2002). The LCT gene is regulated by the nearby MCM6 gene (minichromosome maintenance complex component 6), which encodes a helicase complex. To date, at least four different MCM6 variants have been identified in affecting LCT gene expression (NIH, 2019). Single nucleotide polymorphisms (SNPs) associated with the lactase persistence vary by region. In European populations it is associated with C/T-13910 and G/A-22018 mutations of the LCT gene (Enattah et al., 2002; Hogenauer et al., 2005; Poulter et al., 2003; Ridefelt & Hakansson, 2005). Additional mutations have been identified in Saudi Arabia populations with G-13915 (Imtiaz et al., 2007), and in African tribes with the G-14010, G-13915, and G-13907 polymorphisms (Ingram et al., 2007; Tishkoff et al., 2007). No SNP associated with lactase persistence has been identified in the lactase gene regulatory sequence in Chinese populations (Zheng et al., 2016). In adult patients with homozygous lactase persistence, enzyme levels are 10-times higher than for patients with homozygous non-persistence, and heterozygous individuals (Deng et al., 2015; Enattah et al., 2007).
Lactase deficiency (LD) is defined as markedly reduced brush-border lactase activity relative to the activity observed in infants (Deng et al., 2015). Continued dairy consumption despite low expression of lactase results in unabsorbed lactose being present in the intestinal tract (lactose malabsorption [LM]), which can lead to symptoms of lactose intolerance (LI) in susceptible individuals (Hammer, 2022).
LI is defined by patient reports of abdominal pain, bloating, borborygmi, and diarrhea induced by dairy consumption. Unabsorbed lactose increases the osmotic load thus increasing the intestinal water content, resulting in osmotic diarrhea. Additionally, lactose and other poorly-absorbed oligosaccharides, disaccharides, monosaccharides, and polyols ubiquitous in the diet are readily fermented by the colonic microbiome, leading to production of short-chain fatty acids and gas (mainly hydrogen [H2], carbon dioxide [CO2], and methane [CH4]) (Magge & Lembo, 2012; Shepherd, Lomer, & Gibson, 2013). LI may be associated with nonspecific symptoms, abdominal pain, bloating, flatulence, diarrhea, or vomiting (Hammer, 2022); however, it is unclear whether these symptoms are directly due to lactose ingestion. Although LM is nearly always attributable to LD, it is not possible to make a definitive diagnosis on clinical presentation alone because double-blind trials have shown that the reliability of self-reported LI is very poor (Deng et al., 2015; Suarez, Savaiano, & Levitt, 1995; Zheng et al., 2016).
Determining if reported symptoms of LI are resultant from LD can be approached through several different methods. The gold standard is the measurement of lactase, sucrase, and maltase activity through intestinal biopsies. However, this method is not commonly used due to its invasive nature (Di Rienzo et al., 2013). Other tests, such as the lactose breath test or biochemical blood tests are more frequently used (Furnari et al., 2013; Mattar, Basile-Filho, Kemp, & Santos, 2013; Ponte et al., 2016b). In addition to biochemical blood tests, genetic markers may be useful for LI diagnosis; however, a positive genetic test indicates whether lactase activity decline may represent a clinical problem for the patient, but the test does not give information on actual patient symptoms, making it inappropriate as an initial screening as not all patients with LM will develop symptoms of LI (Ponte et al., 2016a).
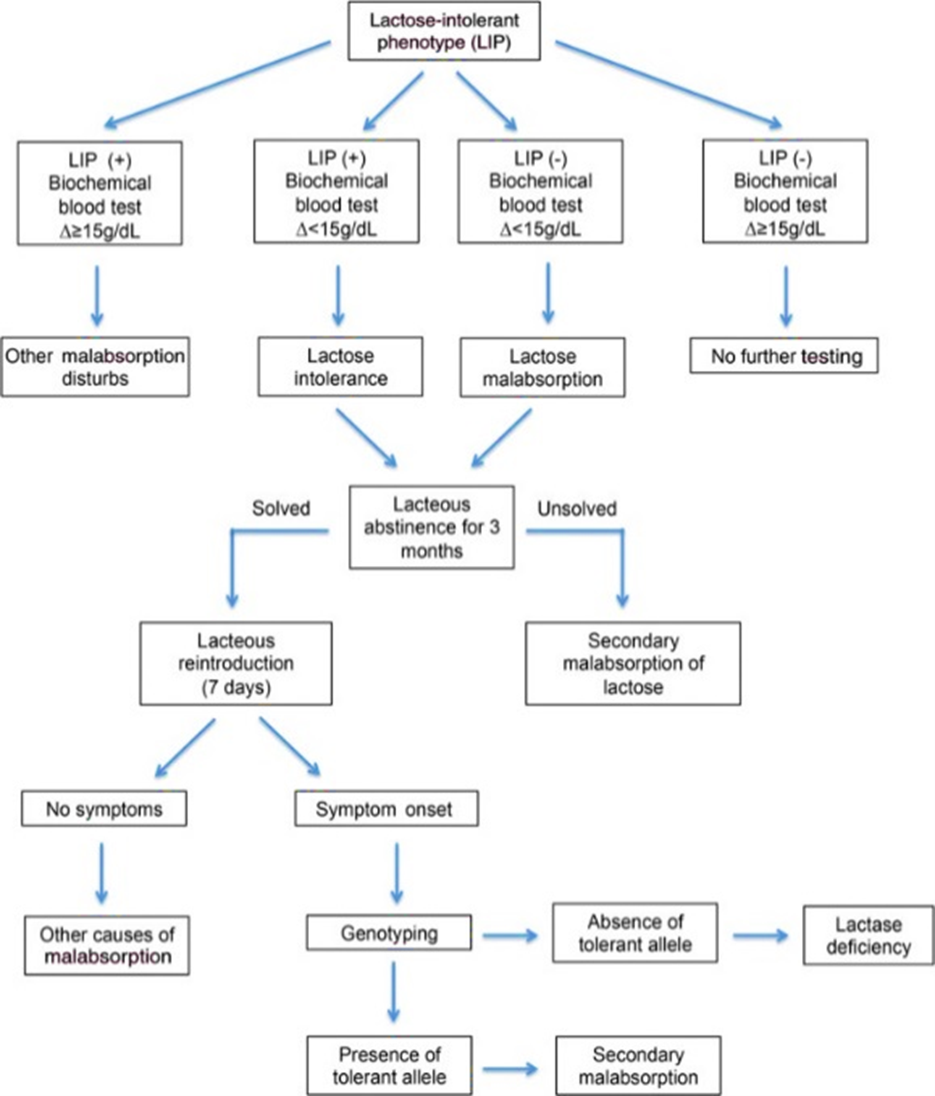
On the contrary, this information is more readily accessible by combining the lactose breath test with intolerance symptom evaluation (Di Stefano et al., 2009). Usual LI management involves excluding milk and milk products from the diet, while ensuring adequate calcium intake (Misselwitz, 2014; Ponte et al., 2016b; Usai-Satta, Scarpa, Oppia, & Cabras, 2012). The use of genetic tests has been proposed as an adjunct to LI diagnosis to differentiate primary LD from secondary causes (Bodlaj et al., 2006) as depicted below in the figure taken from Ponte et al. (2016b).
Clinical Utility and Validity
Marton, Xue, and Szilagyi (2012) compared the common polymorphism C/T 13910 with the lactase breath test and lactose tolerance test to assess each test’s ability to predict genotype/phenotype relationships. The agreement of the breath test and genotype was 0.88 sensitivity and 0.85 specificity whereas the agreement between genotype and tolerance test was 0.94 sensitivity and 0.90 specificity (Marton et al., 2012).
Baffour-Awuah et al. (2015) studied the association of genotypes at the -13910 and -22018 SNPs with clinical characteristics, RNA quantification, and enzymatic phenotypes among a range of European ethnicities within the U.S. population. The authors concluded that “13910T/T genotype will frequently, but not perfectly, predict lactase persistence in this mixed European-ancestry population; a -13910T/C genotype will not predict the phenotype” (Baffour-Awuah et al., 2015).
Misselwitz et al. (2013) stated that genetic testing for the −13910*T genotype in certain African, Arabic, or Asian subpopulations has limited value because lactase persistence may be linked to different polymorphisms. The authors also stated that genetic tests will be negative in patients with secondary causes of lactase deficiency and that no information about clinical symptoms lactose intolerance will be obtained during testing (Misselwitz et al., 2013).
Brasen, Frischknecht, Ornskov, Andreasen, and Madsen (2017) genotyped 3395 routine samples using real-time polymerase chain reaction (PCR) for the -13910C > T-variant to determine the prevalence of the variants in a Danish cohort examined for lactose intolerance as well as to improve the real-time PCR analysis for detection of the different variants. The researchers found that “Using real-time PCR resulted in 100% successful genotyping of the -13910C > T variant. By using a quality value of 99% and sequencing the undetermined samples we improved the ability of the assay to identify variants other than -13910C > T. This resulted in a reduction of the diagnostic error rate by a factor of 2.4 while increasing the expenses only 3%” (Brasen et al., 2017).
Coluccia et al. (2019) compared results from a lactose breath test and a lactase-gene polymorphism test in a total of 158 symptomatic adults to identify lactose intolerance. Whole blood samples were used for genetic testing purposes. Lactose breath testing resulted in positive results in 75.9% of participants, while genetic testing identified lactase-gene polymorphisms in 82.3% of participants. In conclusion, the authors state, “We suggest considering the use of the genetic test after LBT [lactose breath test] administration, when secondary hypolactasia is suspected, for completion of diagnostic procedures” (Coluccia et al., 2019).
Muendlein et al. (2019) developed a real-time PCR protocol for the detection of the LCT-13910C>T variant from whole blood samples. Lactose tolerance in adults is known to be strongly associated with the genetic variant LCT-13910C>T, which lies near the lactose encoding gene. Results from real-time PCR were compared to those from Sanger sequencing. A total of 105 whole blood samples were analyzed. When compared to Sanger sequencing, the real-time PCR protocol was determined to be a reliable method for the detection of the LCT-13910C>T (Muendlein et al., 2019).
In a cross-sectional study by Couce, Sánchez-Pintos, González-Vioque, and Leis (2020), 493 children suspected of functional gastrointestinal disorders (FGID) were studied using the exhaled H2 test, gastrointestinal symptoms, and SNP C/T-13910 genetic testing to observe the correlation between the genotype and phenotype of the C/T-13910 lactase non-persistence (LNP) gene. "The C/T-13910 genotype distribution was as follows: CC, 46.0%; CT, 39.5%; TT 14.4%. The frequency of the LP allele was 34.1%." A significant increase in LNP genotype, H2 test, and gastrointestinal symptoms were observed with increasing age. According to phenotype, CC carriers were mainly lactose non-absorbers (75.4%), TT carriers were lactose-absorbers (91.6%), and CT heterozygotes were predominantly lactose absorbers (90.7%) and lactose tolerant (74.3%). According to these results and its prevalence in older age, the authors state that the C/T-13910 polymorphism is significantly correlated with the phenotype and the authors suggest that the practical value of genetic testing is greater in older children (Couce et al., 2020).
Nardone et al. (2021) studied the prevalence of lactose intolerance assessed by hydrogen breath test (H-BT) and investigated the prevalence of three single genetic polymorphisms of the lactase gene. 54 IBD patients were recruited, and H-BT was positive in 64.8% of IBD patients and 62.3% control patients. The genetic analysis revealed that 46 IBD patients (85.2%) had the wild-type genotype (LCT-13910 CC) while the other polymorphisms (CT-22018, AG-13910, and CT-22018/AG-13910) were less common in IBD patients. In the control group, the wild genotype was found in 87% of the patients. Therefore, the prevalence of polymorphisms did not differ between the IBD group and the control group. The correlation between positive H-BT and genetic analysis showed that the "wild-type genotype was associated with higher rate of lactose intolerance in the total population” (Nardone et al., 2021).
Usai-Satta, Oppia, Lai, and Cabras (2021) reviewed the usefulness of hydrogen breath testing in the nutritional management of carbohydrate digestive disorders, including lactose malabsorption and intolerance, using the online databases Pubmed, Medline, and Cochrane. According to the authors, the literature search yielded that though lactose breath testing is an indirect test for lactose malabsorption, “Lactose BT showed good sensitivity and optimal specificity for lactose malabsorption”, leading them to conclude that “Before starting a low FODMAP [fermentable oligo-, di- and monosaccharides and polyols] diet, lactose BT should be suggested in a population with low prevalence of hypolactasia,” helped in part by its non-invasiveness and inexpensiveness (Usai-Satta et al., 2021).
American Academy of Pediatrics (AAP)
The AAP published guidelines (Heyman, 2006) on the evaluation of Lactose Intolerance in Infants, Children and Adolescents which recommend:
“Children with suspected lactose intolerance can be assessed clinically by dietary lactose elimination or by tests including noninvasive hydrogen breath testing or invasive intestinal biopsy determination of lactase (and other disaccharidase) concentrations. Treatment consists of use of lactase-treated dairy products or oral lactase supplementation, limitation of lactose-containing foods, or dairy elimination. … If dairy products are eliminated, other dietary sources of calcium or calcium supplements need to be provided.”
The AAP also reported that “Recent studies suggest that in the future, genetic testing may be useful for identifying individuals at increased risk of lactase deficiency and consequent diminished bone mineral density, potentially allowing early intervention with dietary manipulation or nutrient supplementation.”
This statement was reaffirmed in 2012.
American College of Gastroenterology (ACG)
The ACG has a webpage focused on lactose intolerance in children. Regarding diagnoses, the ACG states that “Lactose intolerance is diagnosed by a simple test called a hydrogen breath test” (ACG, 2012). Genetic testing was not mentioned.
The 1st Rome Consensus Conference
The 1st Rome Consensus Conference issued their opinion on H2-breath testing for sugar malabsorption. They found that “The determination of lactase activity in jejunal biopsy is currently considered the gold standard for lactose malabsorption. However, its results can be influenced by the irregular dissemination of lactase activity throughout the small intestine mucosa. On the basis of literature review, the lactose breath test is a reliable, non-invasive technique, which is provided with good sensitivity and optimal specificity.”
Though it is acknowledged that there is no unequivocal reference test available for lactose malabsorption testing, the Conference graded the quality of evidence for the above as Class I (“Conditions with evidence or general accord that a particular procedure or treatment is useful or effective”) of Strength A (“Data derived from multiple large and intermediate RCT”) (Gasbarrini et al., 2009).
Regulatory Status
A search for "lactose" on FDA website on May 6, 2021, yielded no results for the genetic testing of lactose intolerance (FDA, 2021). Additionally, many labs have developed specific tests that they must validate and perform in house. These laboratory-developed tests (LDTs) are regulated by the Centers for Medicare & Medicaid Services (CMS) as high-complexity tests under the Clinical Laboratory Improvement Amendments of oo1988 (CLIA '88). As an LDT, the U.S. Food and Drug Administration has not approved or cleared this test; however, FDA clearance or approval is not currently required for clinical use.
References
- ACG. (2012). Lactose Intolerance in Children. Retrieved from https://gi.org/topics/lactose-intolerance-in-children/
- Baffour-Awuah, N. Y., Fleet, S., Montgomery, R. K., Baker, S. S., Butler, J. L., Campbell, C., . . . Hirschhorn, J. N. (2015). Functional significance of single nucleotide polymorphisms in the lactase gene in diverse US patients and evidence for a novel lactase persistence allele at -13909 in those of European ancestry. J Pediatr Gastroenterol Nutr, 60(2), 182-191. doi:10.1097/mpg.0000000000000595
- Bodlaj, G., Stocher, M., Hufnagl, P., Hubmann, R., Biesenbach, G., Stekel, H., & Berg, J. (2006). Genotyping of the lactase-phlorizin hydrolase -13910 polymorphism by LightCycler PCR and implications for the diagnosis of lactose intolerance. Clin Chem, 52(1), 148-151. doi:10.1373/clinchem.2005.057240
- Brasen, C. L., Frischknecht, L., Ornskov, D., Andreasen, L., & Madsen, J. S. (2017). Combination of real-time PCR and sequencing to detect multiple clinically relevant genetic variations in the lactase gene. Scand J Clin Lab Invest, 77(1), 60-65. doi:10.1080/00365513.2016.1261408
- Coluccia, E., Iardino, P., Pappalardo, D., Brigida, A. L., Formicola, V., De Felice, B., . . . De Magistris, L. (2019). Congruency of Genetic Predisposition to Lactase Persistence and Lactose Breath Test. Nutrients, 11(6). doi:10.3390/nu11061383
- Couce, M. L., Sánchez-Pintos, P., González-Vioque, E., & Leis, R. (2020). Clinical Utility of LCT Genotyping in Children with Suspected Functional Gastrointestinal Disorder. Nutrients, 12(10), 3017. Retrieved from https://www.mdpi.com/2072-6643/12/10/3017
- Deng, Y., Misselwitz, B., Dai, N., & Fox, M. (2015). Lactose Intolerance in Adults: Biological Mechanism and Dietary Management. Nutrients, 7(9), 8020-8035. doi:10.3390/nu7095380
- Di Rienzo, T., D'Angelo, G., D'Aversa, F., Campanale, M. C., Cesario, V., Montalto, M., . . . Ojetti, V. (2013). Lactose intolerance: from diagnosis to correct management. Eur Rev Med Pharmacol Sci, 17 Suppl 2, 18-25. Retrieved from https://www.europeanreview.org/wp/wp-content/uploads/018-025.pdf
- Di Stefano, M., Terulla, V., Tana, P., Mazzocchi, S., Romero, E., & Corazza, G. R. (2009). Genetic test for lactase non-persistence and hydrogen breath test: is genotype better than phenotype to diagnose lactose malabsorption? Dig Liver Dis, 41(7), 474-479. doi:10.1016/j.dld.2008.09.020
- Enattah, N. S., Kuokkanen, M., Forsblom, C., Natah, S., Oksanen, A., Jarvela, I., . . . Savilahti, E. (2007). Correlation of intestinal disaccharidase activities with the C/T-13910 variant and age. World J Gastroenterol, 13(25), 3508-3512. Retrieved from http://dx.doi.org/
- Enattah, N. S., Sahi, T., Savilahti, E., Terwilliger, J. D., Peltonen, L., & Jarvela, I. (2002). Identification of a variant associated with adult-type hypolactasia. Nat Genet, 30(2), 233-237. doi:10.1038/ng826
- Furnari, M., Bonfanti, D., Parodi, A., Franze, J., Savarino, E., Bruzzone, L., . . . Savarino, V. (2013). A comparison between lactose breath test and quick test on duodenal biopsies for diagnosing lactase deficiency in patients with self-reported lactose intolerance. J Clin Gastroenterol, 47(2), 148-152. doi:10.1097/MCG.0b013e31824e9132
- Gasbarrini, A., Corazza, G. R., Gasbarrini, G., Montalto, M., Di Stefano, M., Basilisco, G., . . . Tursi, A. (2009). Methodology and indications of H2-breath testing in gastrointestinal diseases: the Rome Consensus Conference. Aliment Pharmacol Ther, 29 Suppl 1, 1-49. doi:10.1111/j.1365-2036.2009.03951.x
- Hammer, H. F., Hogenauer, Christoph. (2022). Lactose intolerance and malabsorption: Clinical manifestations, diagnosis, and management. In S. Grover (Ed.), UpToDate. Retrieved from https://www.uptodate.com/contents/lactose-intolerance-clinical-manifestations-diagnosis-and-management?search=lactose%20intolerance&source=search_result&selectedTitle=1~150&usage_type=default&display_rank=1
- Heyman, M. B. (2006). Lactose intolerance in infants, children, and adolescents. Pediatrics, 118(3), 1279-1286. doi:10.1542/peds.2006-1721
- Hogenauer, C., Hammer, H. F., Mellitzer, K., Renner, W., Krejs, G. J., & Toplak, H. (2005). Evaluation of a new DNA test compared with the lactose hydrogen breath test for the diagnosis of lactase non-persistence. Eur J Gastroenterol Hepatol, 17(3), 371-376. Retrieved from http://dx.doi.org/
- Imtiaz, F., Savilahti, E., Sarnesto, A., Trabzuni, D., Al-Kahtani, K., Kagevi, I., . . . Jarvela, I. (2007). The T/G 13915 variant upstream of the lactase gene (LCT) is the founder allele of lactase persistence in an urban Saudi population. J Med Genet, 44(10), e89. doi:10.1136/jmg.2007.051631
- Ingram, C. J., Elamin, M. F., Mulcare, C. A., Weale, M. E., Tarekegn, A., Raga, T. O., . . . Swallow, D. M. (2007). A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum Genet, 120(6), 779-788. doi:10.1007/s00439-006-0291-1
- Luyt, D., Ball, H., Makwana, N., Green, M. R., Bravin, K., Nasser, S. M., & Clark, A. T. (2014). BSACI guideline for the diagnosis and management of cow's milk allergy. Clinical & Experimental Allergy, 44(5), 642-672. doi:10.1111/cea.12302
- Magge, S., & Lembo, A. (2012). Low-FODMAP Diet for Treatment of Irritable Bowel Syndrome. Gastroenterol Hepatol (N Y), 8(11), 739-745. Retrieved from http://dx.doi.org/
- Marton, A., Xue, X., & Szilagyi, A. (2012). Meta-analysis: the diagnostic accuracy of lactose breath hydrogen or lactose tolerance tests for predicting the North European lactase polymorphism C/T-13910. Aliment Pharmacol Ther, 35(4), 429-440. doi:10.1111/j.1365-2036.2011.04962.x
- Mattar, R., Basile-Filho, A., Kemp, R., & Santos, J. S. (2013). Comparison of Quick Lactose Intolerance Test in duodenal biopsies of dyspeptic patients with single nucleotide polymorphism LCT-13910C>T associated with primary hypolactasia/lactase-persistence. Acta Cir Bras, 28 Suppl 1, 77-82. Retrieved from http://dx.doi.org/
- Misselwitz, B. (2014). Lactose intolerance: new insights due to blinded testing? Digestion, 90(1), 72-73. doi:10.1159/000365144
- Misselwitz, B., Pohl, D., Fruhauf, H., Fried, M., Vavricka, S. R., & Fox, M. (2013). Lactose malabsorption and intolerance: pathogenesis, diagnosis and treatment. United European Gastroenterol J, 1(3), 151-159. doi:10.1177/2050640613484463
- Muendlein, A., Leiherer, A., Zach, C., Brandtner, E. M., Fraunberger, P., Drexel, H., & Geiger, K. (2019). Real-time PCR based detection of the lactase non-persistence associated genetic variant LCT-13910C>T directly from whole blood. Mol Biol Rep, 46(2), 2379-2385. doi:10.1007/s11033-019-04696-9
- Nardone, O. M., Manfellotto, F., D’Onofrio, C., Rocco, A., Annona, G., Sasso, F., . . . Castiglione, F. (2021). Lactose Intolerance Assessed by Analysis of Genetic Polymorphism, Breath Test and Symptoms in Patients with Inflammatory Bowel Disease. Nutrients, 13(4), 1290. Retrieved from https://www.mdpi.com/2072-6643/13/4/1290
- NIH. (2019, 03/19/2019). MCM6 gene. Genetics Home Reference. Retrieved from https://ghr.nlm.nih.gov/gene/MCM6
- Ponte, P. R., de Medeiros, P. H., Havt, A., Caetano, J. A., Cid, D. A., Prata Mde, M., . . . Lima, A. (2016a). Clinical evaluation, biochemistry and genetic polymorphism analysis for the diagnosis of lactose intolerance in a population from northeastern Brazil. Clinics (Sao Paulo), 71(2), 82-89. doi:10.6061/clinics/2016(02)06
- Ponte, P. R., de Medeiros, P. H., Havt, A., Caetano, J. A., Cid, D. A., Prata Mde, M., . . . Lima, A. A. (2016b). Clinical evaluation, biochemistry and genetic polymorphism analysis for the diagnosis of lactose intolerance in a population from northeastern Brazil. Clinics (Sao Paulo), 71(2), 82-89. doi:10.6061/clinics/2016(02)06
- Poulter, M., Hollox, E., Harvey, C. B., Mulcare, C., Peuhkuri, K., Kajander, K., . . . Swallow, D. M. (2003). The causal element for the lactase persistence/non-persistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann Hum Genet, 67(Pt 4), 298-311. Retrieved from http://dx.doi.org/
- Ridefelt, P., & Hakansson, L. D. (2005). Lactose intolerance: lactose tolerance test versus genotyping. Scand J Gastroenterol, 40(7), 822-826. doi:10.1080/00365520510015764
- Shepherd, S. J., Lomer, M. C., & Gibson, P. R. (2013). Short-chain carbohydrates and functional gastrointestinal disorders. Am J Gastroenterol, 108(5), 707-717. doi:10.1038/ajg.2013.96
- Suarez, F. L., Savaiano, D. A., & Levitt, M. D. (1995). A comparison of symptoms after the consumption of milk or lactose-hydrolyzed milk by people with self-reported severe lactose intolerance. N Engl J Med, 333(1), 1-4. doi:10.1056/nejm199507063330101
- Swallow, D. M. (2003). Genetics of lactase persistence and lactose intolerance. Annu Rev Genet, 37, 197-219. doi:10.1146/annurev.genet.37.110801.143820
- Tishkoff, S. A., Reed, F. A., Ranciaro, A., Voight, B. F., Babbitt, C. C., Silverman, J. S., . . . Deloukas, P. (2007). Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet, 39(1), 31-40. doi:10.1038/ng1946
- Usai-Satta, P., Oppia, F., Lai, M., & Cabras, F. (2021). Hydrogen Breath Tests: Are They Really Useful in the Nutritional Management of Digestive Disease? Nutrients, 13(3). doi:10.3390/nu13030974
- Usai-Satta, P., Scarpa, M., Oppia, F., & Cabras, F. (2012). Lactose malabsorption and intolerance: What should be the best clinical management? World J Gastrointest Pharmacol Ther, 3(3), 29-33. doi:10.4292/wjgpt.v3.i3.29
- Wang, Y., Harvey, C. B., Pratt, W. S., Sams, V. R., Sarner, M., Rossi, M., . . . Swallow, D. M. (1995). The lactase persistence/non-persistence polymorphism is controlled by a cis-acting element. Hum Mol Genet, 4(4), 657-662. Retrieved from http://dx.doi.org/
- Zheng, X., China, Z. U. D. o. G. H., Chu, H., China, Z. U. D. o. G. H., Cong, Y., China, Z. U. D. o. G. H., . . . Switzerland, U. o. Z. Z. C. f. I. H. P. Z. Z. (2016). Self‐reported lactose intolerance in clinic patients with functional gastrointestinal symptoms: prevalence, risk factors, and impact on food choices. Neurogastroenterology & Motility, 27(8), 1138-1146. doi:10.1111/nmo.12602
Coding Section
| Codes | Number | Description |
| CPT | 81400 |
Molecular pathology procedure, level 1 (e.g., identification of single germline variant [e.g., SNP] by techniques such as restriction enzyme digestion or melt curve analysis) includes the following test effective 07/01/13: LCT (lactase-phlorizin hydrolase) (e.g., lactose intolerance), 13910 C>T variant |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community, Blue Cross Blue Shield Association technology assessment program (TEC) and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies, and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2014 Forward
|
07/19/2022 |
Annual review, no change to policy intent. Updated description, rationale and references |
|
07/21/2021 |
Annual review, no change to policy intent. Updating rationale and references. Removing regulatory status as that information is included in the rationale. |
|
07/14/2020 |
Annual review, no change to policy intent. Reformatting entire policy for clarity. |
|
07/12/2019 |
Annual review, adding :2. Genetic testing of the LCT gene and/or MCM6 gene for lactose intolerance and/or lactase insufficiency is considered INVESTIGATIONAL to address proprietary genetic tests. |
|
07/18/2018 |
Annual review, no change to policy intent. |
|
07/12/2017 |
Annual review, no change to policy intent. |
|
04/25/2017 |
Updated category to Laboratory. No other changes |
|
06/01/2016 |
Annual review, no change to policy intent. |
|
06/22/2015 |
Annual review, no change to policy intent. Updated background, description, rationale and references. Added coding, guidelines and appendix 1. |
|
06/12/2014 |
Annual review, no changes made. |